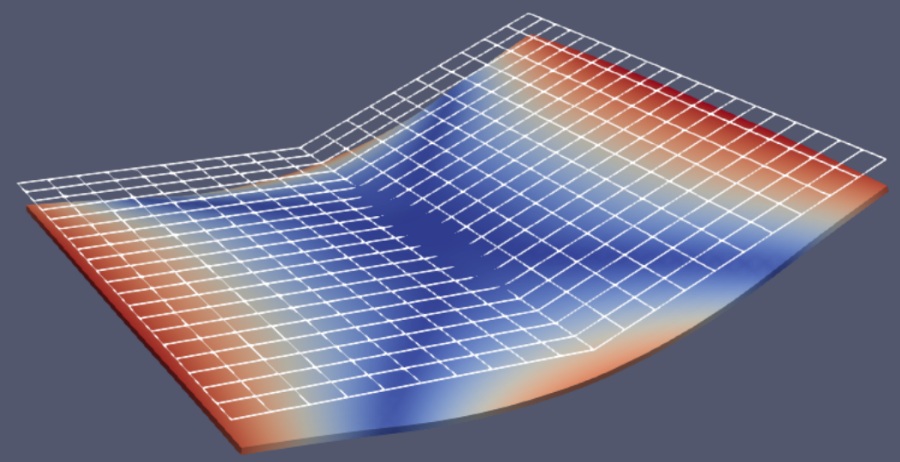
Case study III: liquid crystal elastomer shape matching¶
Our goals is to design the ordered regions and the director angles on the ordered regions of a liquid crystal elastomer film with four layers, and the director angles on the regions to match a target shape of a 2.5 degree folding:
The variational form for the problem is
\[\int_{\Omega} \sigma:\nabla v d x =0,\]
where the \(\sigma\), \(v\) are the stress tenser and the test functions.
The code can be downloaded from here
1. Code¶
We explain the code in detail in this section.
1.1. Import¶
First, import dolfin, meshio, numpy, pygmsh, scipy
atomics.api, atomics.pde, and atomics.general_filter_comp
, as well as the stock OpenMDAO components such as extract_comp, and interpolant_comp.
import dolfin as df
import meshio
import numpy as np
import pygmsh
import scipy.sparse
from scipy import spatial
import openmdao.api as om
from atomics.api import PDEProblem, AtomicsGroup
from atomics.pdes.thermo_mechanical_lce import get_residual_form
from atomics.general_filter_comp import GeneralFilterComp
from atomics.extract_comp import ExtractComp
from atomics.interpolant_comp import InterpolantComp
from atomics.copy_comp import Copycomp
from atomics.symmetric_angle_comp import SymmericAnglecomp
from atomics.symmetric_rho_comp import SymmericRhocomp
'''
code for LCE topology optimization
'''
'''
Define constants
'''
# parameters for the film
LENGTH = 2.5e-3
WIDTH = 5e-3
THICKNESS = 5e-5
START_X = -2.5e-3
START_Y = -2.5e-3
START_Z = -2.5e-6
NUM_ELEMENTS_X = NUM_ELEMENTS_Y = 50
NUM_ELEMENTS_Z = 4
K = df.Constant(5.e6)
ALPHA = 2.5e-3
degree = 2.5
angle = np.pi/180 * degree
# angle = 0
1.2. Define the mesh¶
mesh = df.BoxMesh.create(
[df.Point(-LENGTH, -WIDTH/2, 0), df.Point(LENGTH, WIDTH/2, THICKNESS)],
[NUM_ELEMENTS_X, NUM_ELEMENTS_Y, NUM_ELEMENTS_Z],
df.CellType.Type.hexahedron,
)
'''
Define bcs (middle lines to preserve symmetry)
'''
class MidHBoundary(df.SubDomain):
def inside(self, x, on_boundary):
return ( abs(x[1] - (START_Y-START_Y)) < df.DOLFIN_EPS )
class MidVBoundary(df.SubDomain):
def inside(self, x, on_boundary):
return ( abs(x[0] - (START_X-START_X)) < df.DOLFIN_EPS)
class MidZBoundary(df.SubDomain):
def inside(self, x, on_boundary):
return ( abs(x[0] - (START_X-START_X)) < df.DOLFIN_EPS *1e12
and abs(x[2] + 0) < df.DOLFIN_EPS *1e9
and abs(x[1] - (START_Y-START_Y)) < df.DOLFIN_EPS*1e12)
1.3. Define the PDE problem¶
# PDE problem
pde_problem = PDEProblem(mesh)
'''
Add input to the PDE problem
'''
# name = 'density', function = density_function
density_function_space = df.FunctionSpace(mesh, 'DG', 0)
density_function = df.Function(density_function_space)
pde_problem.add_input('density', density_function)
# name = 'angle', function = angle_function
angle_function_space = df.FunctionSpace(mesh, 'DG', 0)
angle_function = df.Function(angle_function_space)
pde_problem.add_input('angle', angle_function)
'''
Add states
'''
# Define displacements function
displacements_function_space = df.VectorFunctionSpace(mesh, 'Lagrange', 1)
displacements_function = df.Function(displacements_function_space)
v = df.TestFunction(displacements_function_space)
residual_form = get_residual_form(
displacements_function,
v,
density_function,
angle_function,
K,
ALPHA
)
pde_problem.add_state('displacements', displacements_function, residual_form, 'density', 'angle')
'''
Add output
'''
# Add output-avg_density to the PDE problem:
volume = df.assemble(df.Constant(1.) * df.dx(domain=mesh))
avg_density_form = density_function / (df.Constant(1. * volume)) * df.dx(domain=mesh)
pde_problem.add_scalar_output('avg_density', avg_density_form, 'density')
# Add output-errorL2 to the PDE problem:
desired_disp = df.Expression(( "-(1-cos(angle))*x[0]",
"0.0",
"abs(x[0])*sin(angle)"),
angle=angle,
degree=1 )
# desired_disp = df.project(desired_disp, displacements_function_space )
vol = df.assemble(df.Constant(1) *df.dx(domain=mesh))
e = desired_disp - displacements_function
norm_form = e**2/vol*df.Constant(1e9)*df.dx(domain=mesh)
# norm = df.assemble(e**2/vol*df.dx(domain=mesh))
pde_problem.add_scalar_output('error_norm', norm_form, 'displacements')
'''
4. 3. Add bcs
'''
bc_displacements_v = df.DirichletBC(displacements_function_space.sub(0),
df.Constant((0.0)),
MidVBoundary())
bc_displacements_h = df.DirichletBC(displacements_function_space.sub(1),
df.Constant((0.0)),
MidHBoundary())
bc_displacements_z = df.DirichletBC(displacements_function_space.sub(2),
df.Constant((0.0)),
MidZBoundary())
# Add boundary conditions to the PDE problem:
pde_problem.add_bc(bc_displacements_v)
pde_problem.add_bc(bc_displacements_h)
pde_problem.add_bc(bc_displacements_z)
1.4. Set up the OpenMDAO model¶
'''
Add OpenMDAO comps & groups
'''
# Define the OpenMDAO problem and model
prob = om.Problem()
num_dof_density = pde_problem.inputs_dict['density']['function'].function_space().dim()
bot_idx = np.arange(int(num_dof_density/4/4))
top_idx = np.arange(int(num_dof_density/2/4)-int(num_dof_density/4/4), int(num_dof_density/2/4))
ini_angle = np.zeros(int(num_dof_density/2/4))
ini_angle[bot_idx] = np.pi/2
# Add IndepVarComp-density_unfiltered & angle
comp = om.IndepVarComp()
comp.add_output(
'density_unfiltered_layer_q',
shape=int(density_function_space.dim()/4/4),
val=np.ones((int(density_function_space.dim()/4/4))),
)
# comp.add_output(
# 'density_unfiltered',
# shape=int(density_function_space.dim()),
# val=np.ones(int(density_function_space.dim())),
# )
comp.add_output(
'angle_t_b_q',
shape=ini_angle.shape,
val=ini_angle, #TO be fixed
# val=np.random.random(num_dof_density) * 0.86,
)
prob.model.add_subsystem('indep_var_comp', comp, promotes=['*'])
print('indep_var_comp')
comp = SymmericRhocomp(
in_name='density_unfiltered_layer_q',
out_name='density_unfiltered_layer',
in_shape=int(density_function_space.dim()/16),
num_copies = 4,
)
prob.model.add_subsystem('sym_rho_comp', comp, promotes=['*'])
comp = SymmericAnglecomp(
in_name='angle_t_b_q',
out_name='angle_t_b',
in_shape=int(ini_angle.size),
num_copies = 4,
)
prob.model.add_subsystem('sym_angle_comp', comp, promotes=['*'])
# add copy comp
comp = Copycomp(
in_name='density_unfiltered_layer',
out_name='density_unfiltered',
in_shape=int(density_function_space.dim()/4),
num_copies = 4,
)
prob.model.add_subsystem('copy_comp', comp, promotes=['*'])
# Add interpolant
comp = InterpolantComp(
in_name='angle_t_b',
out_name='angle',
in_shape=int(density_function_space.dim()/2),
num_pts = 4,
)
prob.model.add_subsystem('interpolant_comp', comp, promotes=['*'])
# Add filter
comp = GeneralFilterComp(density_function_space=density_function_space)
prob.model.add_subsystem('general_filter_comp', comp, promotes=['*'])
# Add AtomicsGroup
group = AtomicsGroup(pde_problem=pde_problem)
prob.model.add_subsystem('atomics_group', group, promotes=['*'])
# prob.model.add_design_var('density_unfiltered',upper=1., lower=1e-4)
prob.model.add_design_var('density_unfiltered_layer_q',upper=1., lower=1e-4)
prob.model.add_design_var('angle_t_b_q', upper=np.pi, lower=0.)
prob.model.add_objective('error_norm')
prob.model.add_constraint('avg_density',upper=0.4, linear=True)
prob.driver = driver = om.pyOptSparseDriver()
driver.options['optimizer'] = 'SNOPT'
driver.opt_settings['Verify level'] = 0
driver.opt_settings['Major iterations limit'] = 7000
driver.opt_settings['Minor iterations limit'] = 1000000
driver.opt_settings['Iterations limit'] = 100000000
driver.opt_settings['Major step limit'] = 2.0
driver.opt_settings['Major feasibility tolerance'] = 1.0e-5
driver.opt_settings['Major optimality tolerance'] =1.e-7
prob.setup()
# prob.check_partials(compact_print=True)
# print(prob['compliance']); exit()
prob.run_driver()
#save the solution vector
df.File('solutions/case_3/lce/displacements.pvd') << displacements_function
df.File('solutions/case_3/lce/angles.pvd') << angle_function
stiffness = df.project(density_function/(1 + 8. * (1. - density_function)), density_function_space)
df.File('solutions/case_3/lce/stiffness.pvd') << stiffness
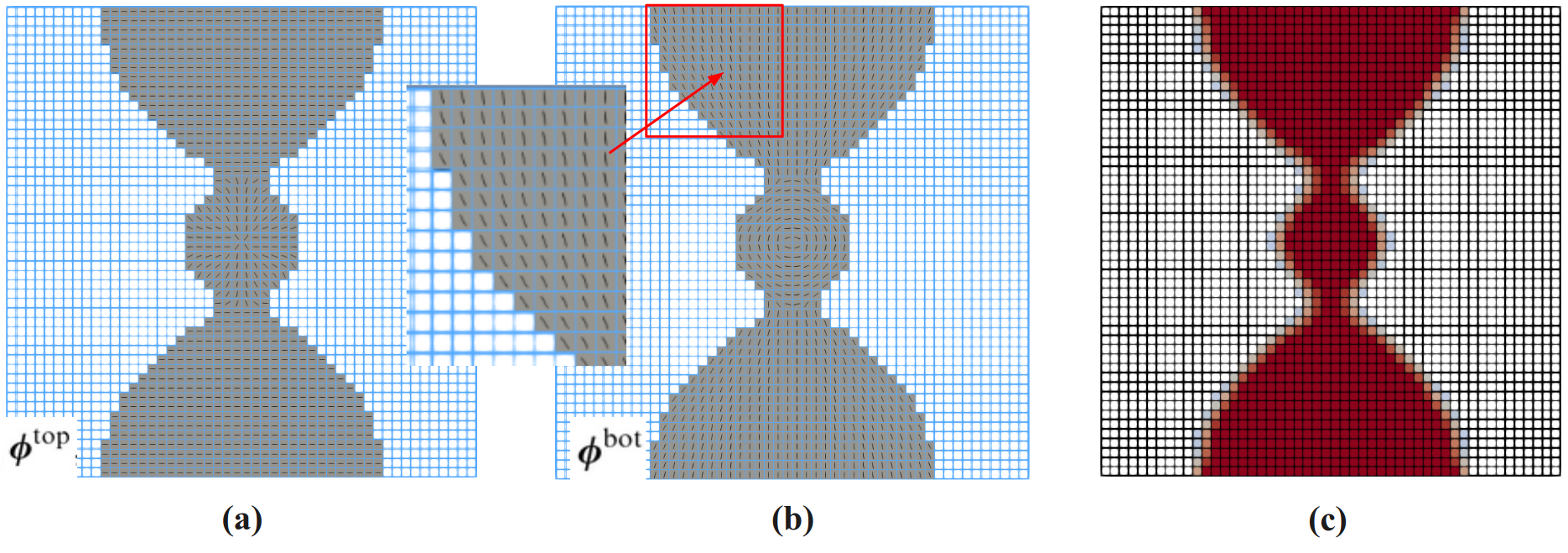
2. Results (density and angle plots)¶
The users can visualize the optimized densities by opening the stiffness.pvd from Paraview.